Hierarchical morphology of organisms, tissues and biomaterials in relation to genetics and environment
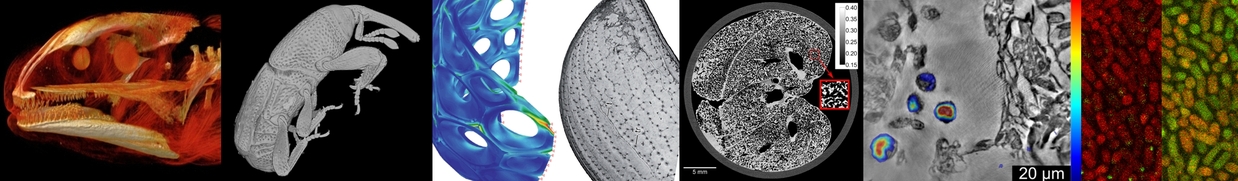
Imaging of different hierarchical levels, such as entire organisms, organs, tissues, cellular networks, and down to single cells, is a central challenge in various fields of biology. Advanced synchrotron X-ray imaging enables digitization of morphology up to complete morphological atlases and phenotype data bases. Morphological data, in correlation with molecular biology and genetic data, has the potential to further enhance our understanding of biological processes in dependence of the genome and external influences (such as pharmaceuticals, biocompatible materials, or other environmental influences). Supported by the BMBF projects X-REGIO, CODE-VITA, ASTOR, and NOVA, an interdisciplinary group of physicists, biologists, and computer scientists of IPS and LAS contribute to the overall goal to relate structure to properties and functionality by combining CL/CT and microscopy for multi-scale and multiple contrasts into hierarchical and correlated imaging modalities, and by comparing with molecular biology and other techniques of our collaboration partners. Further, we apply 4D in vitro & in vivo imaging to study biological processes in real time, such as morpho-dynamics as well as cellular movements. We mainly focus on 3D & 4D phenotype imaging of vertebrate models for developmental biology and genetics, and on morphology and morpho-dynamics of arthropods, including paleontology and biomimetics. Supported by the BMBF X-REGIO and IntelBio-Comp projects, we recently started to study in addition the structure and biocompatibility of scaffolds for tissue engineering (tissue and cell adhesion, proliferation and differentiation) [1-4].
A common downside of established high-resolution biological imaging techniques like light-sheet microscopy, multi-photon microscopy, or electron microscopy is that specimens must be transparent or of relatively small dimensions. Due to these limitations, mechanical sectioning of organs or tissues is required for imaging. Recent developments in X-ray imaging techniques provide us a new way of looking at small organisms. Penetrating thick and opaque tissues, and taking benefit from a relatively large beam size at our imaging stations at KIT, we are able to provide a complete tomographic view even within cm-sized specimens. We complement the studies by multiple experiments at low emittance facilities such as ESRF, the Taiwan Photon Source, and APS. Different protocols provide high-resolution 3D data of entire animals by imparting various X-ray contrasts to soft tissue, otherwise transparent to X-rays, and new algorithms of semi-automated segmentation. Especially the established ability of high sample-throughput and (semi-)automated segmentation paves the way for correlated phenotype screening / genome sequencing.
3D & 4D phenotype imaging of vertebrate models: These activities have been performed in collaboration with the University of Heidelberg by employing high-throughput imaging, hierarchical & correlated imaging, as well as 4D in vivo imaging, aiming to study zebrafish, medaka, and the embyos of the South African clawed frog Xenopus laevis. They are important vertebrate model organisms for studies ranging from developmental biology, over functional genomics, toxicology etc. up to drug development, tissue engineering, cancer research, anesthesia, and other preclinical studies. The understanding of developmental processes and functions of genes in model organisms requires molecular biological approaches in combination with imaging studies of the tissue and organ formation in wild type organisms, variable phenotypes and mutants, both postmortem and in vivo. While a number of technological advances has offered novel approaches to biological imaging with light microscopy no methods hitherto existed for imaging of adult model organisms (e.g., zebrafish, medaka) or optically opaque embryos of Xenopus laevis.
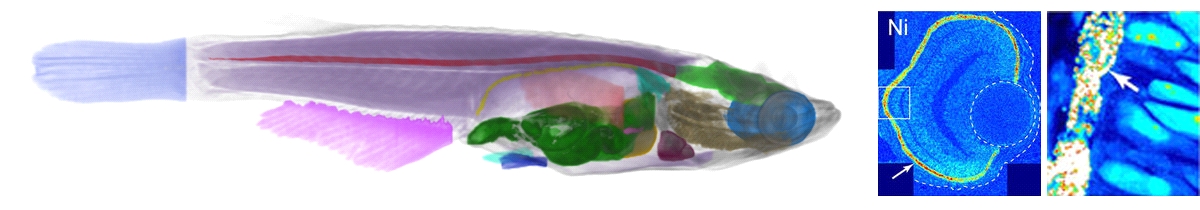
We have performed full animal CT of medaka, aiming to understand stem-cell progression in vertebrates, in particular to address whether the mode of stem cell seperation follows socalled phylogenetic or ontogenetic routes [5]. Recently, tissue imaging protocols for medaka and zebrafish models have been improved for high-throughput screening to study typical phenotype characters of various inbred lines. We are further challenging a new way of looking at gene expression and aiming to develop a general, automated high-throughput method of showing complete 3D expression data of genes with a resolution down to the subcellular level, achievable even in large specimen up to complete animals without the need for any sectioning or mechanical manipulation. In first proof of principle experiments, we could recently visualize gene expression as a new imaging modality on GFP-tagged reporter lines of medaka. Correlating label-free quantitative spatially resolved elemental analysis via trace element distributions over a hierarchy of length scales with further molecular-biological methods, we could study melanosomes in pigmented epithelia maintaining eye lens transparency during zebrafish embryonic development, where oxidative stress has been proposed to be involved in eye diseases, including cataract formation. Comparing knock-down mutants and wild-type representatives of zebrafish we found that the knock-down process eliminates the accumulation of these elements in the retinal pigment epithelium (RPE), indicating that they are bound by mature melanosomes [6].
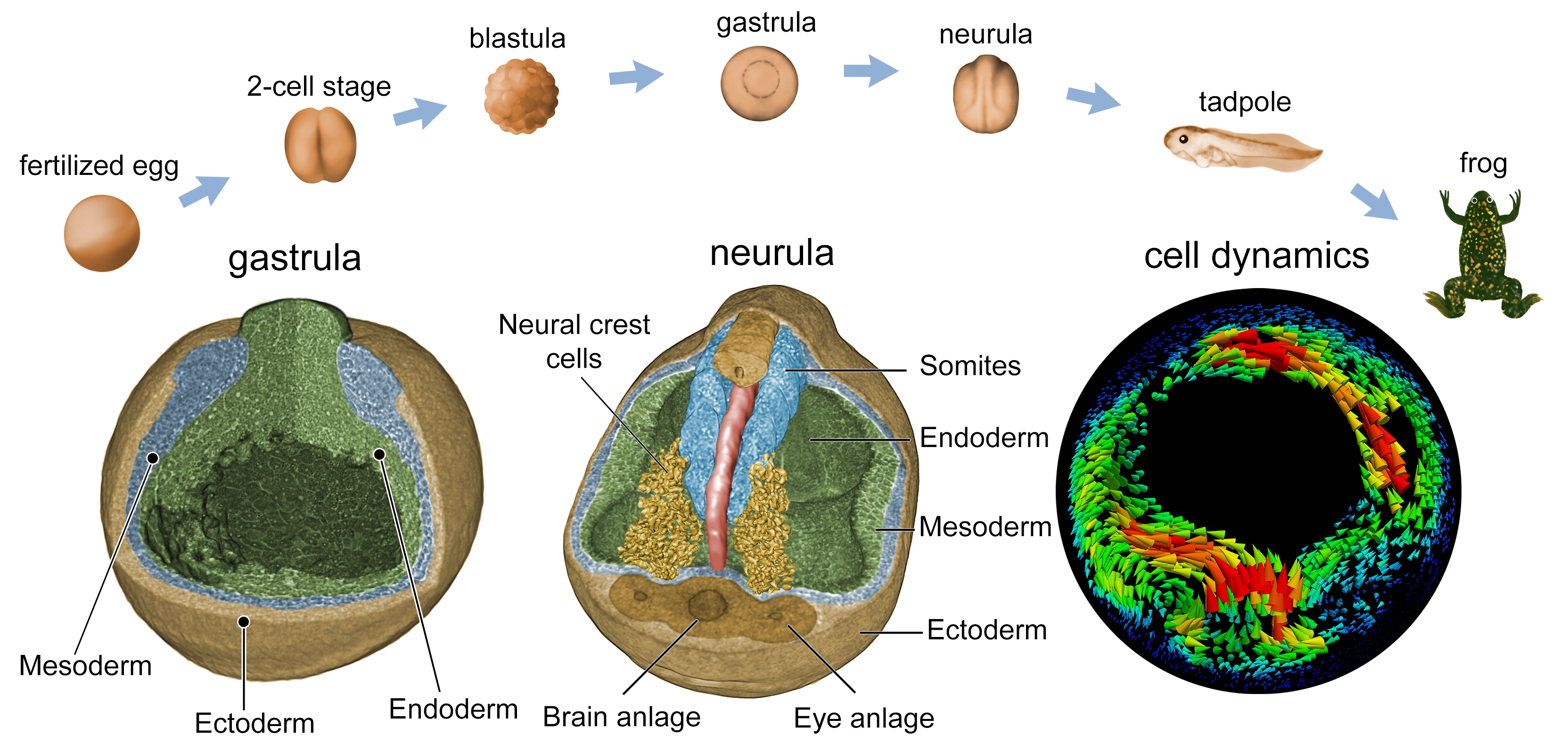
Based on advances in single distance phase reconstruction and optical flow algorithms [7], IPS and LAS were able to perform time-lapsed in vivo imaging, visualizing the 4D spatiotemporal movement of cells and cell groups during the gastrulation stage of Xenopus embryonic development. In all those stages, collective versus differential tissue and cell movements can be observed online with sub-cellular spatiotemporal resolution [8-10]. Further dose optimization allowed us to extend the investigation to later developmental stages, such as neurulation.
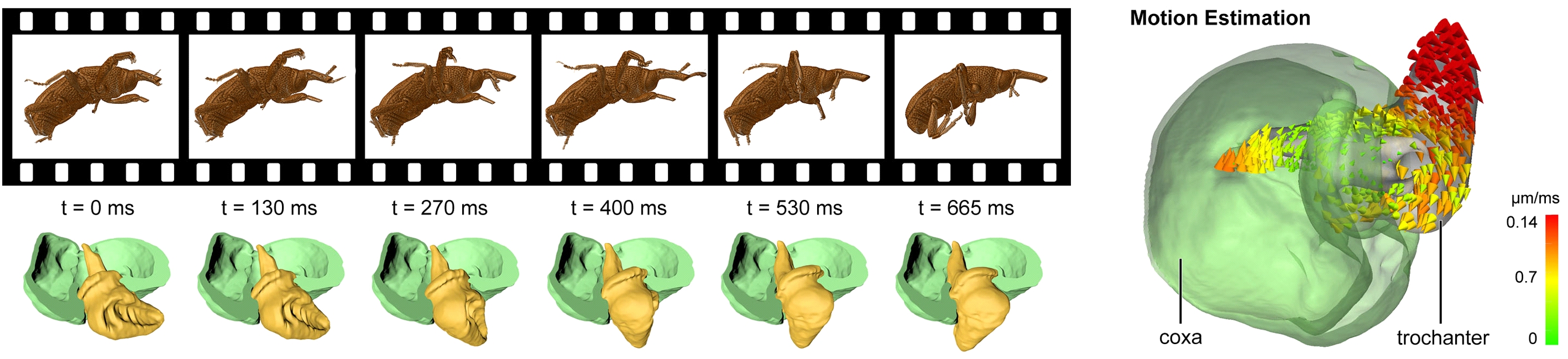
Zoology – from morphology to morpho-dynamics: UFO has become an important tool for gaining complete internal and external morphological information of arthropods. Combination of postmortem and in vivo imaging allowed extending morphology studies to morphodynamics. After our discovery of the first biological screw forming a hip-joint of the Papuan weevil Trigonopterus oblongus [11], we performed a detailed analysis of morpho-dynamical behavior of such hip-joints comparing various species. Cine-tomography enabled real-time study of the physiology by tracking the 4D morphological dynamics of minute anatomical features. Reconstructing the complete 4D spatio-temporal kinematics of a fast-moving screw-and-nut type hip joint, the technique has proven to be a promising new tool to such studies in millimeter-sized animals [12]. UFO realizes the fastest reported CT sequences for sub-10 µm resolution. We provided a method to zoologists to create interactive 3D reconstructions to investigate functional morphology. Animated 3D models combine advantages of movie and interactivity, suitable to simulate largely deterministic movements [13]. In another project, X-ray videos have revealed internal morphological dynamics during mating of bush crickets [14].
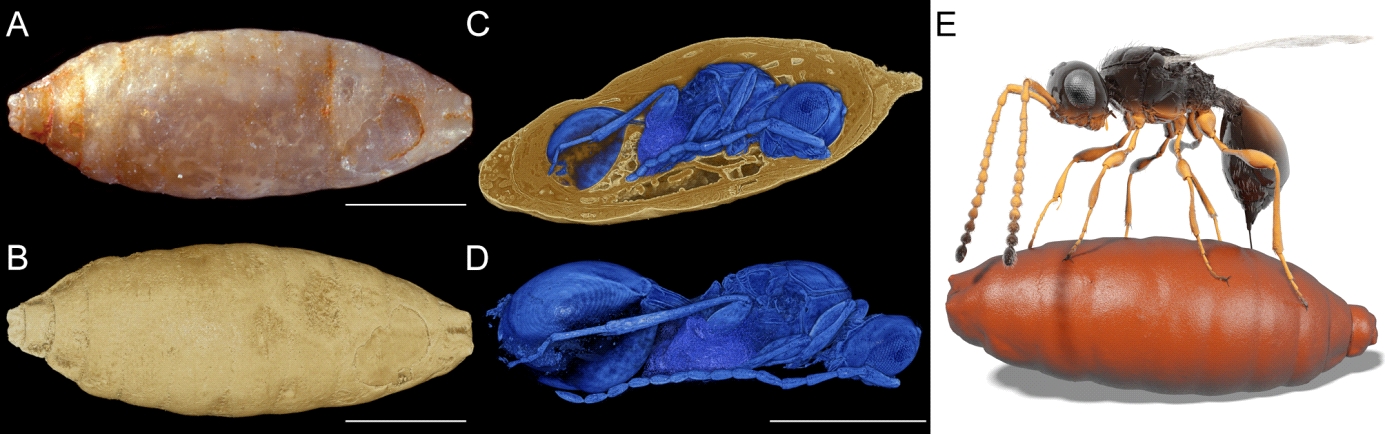
Paleontology – CT of phosphatized arthropods from fissure fillings: Arthropod studies have been extended to fossils which, to date, were largely restricted to external morphology, lacking information about internal characters that are crucial to establish systematic position, ecological role, and evolutionary trends. Moreover, the fossil record of arthropods containing internal characters was dominated by amber inclusions, which show representational bias. By high throughput CT, we have discovered well-preserved 3D anatomy in 30-million-year old mineralized beetles from the fissure fillings in Quercy (south-central France), facilitating a detailed description of the species and its systematic relationship [15]. Despite being known for over a century, these fossils have been neglected, mainly because of the poor condition of their outer surfaces. Our results now provide an incredibly detailed look at their internal structures, demonstrating that mineralized fossils, even those of apparently poor preservation, constitute a rich but yet largely unexploited source of anatomical data of fossil arthropods. By scanning thousands of samples of the fossil fly puparia (Natural History Museums of Basel and Stockholm) from the same locality, we discovered 55 parasitic wasps that belong to four different species. Three of these were preserved with both sexes. Our results allow reconstructing an ancient host-parasite complex by analyzing the biological, morphological, and ecological information [16]. It was thereby also demonstrated that UFO is capable to processing huge amounts of biological data by providing 3D anatomical information even of fully concealed specimens.
Biomimetics: Being natural lightweight constructions, elytra of beetles constitute promising role models for biomimetic development. Based on our µCT-data, partners of IPS & LAS have constructed a research pavilion in order to evaluate fiber composites for architecture and to develop fabrication methods for fiber-reinforced polymer structures. Functional principles of the elytra of the Colorado potato beetle (Leptinotarsa decemlineata) and the Green tortoise beetle (Cassida viridis) were transferred into the modular pavilion.

Related publications
-
B. Göppert, T. Sollich, P. Abaffy, A. Cecilia, J. Heckmann, et al., Superporous Poly (ethylene glycol) Diacrylate Cryogel with a defined elastic modulus for prostate cancer cell research, Small 12 (2016) 3985–3994.
-
Cecilia, A. Baecker, E. Hamann, A. Rack, T. van de Kamp, et al., Optimizing structural and mechanical properties of cryogel scaffolds for use in prostate cancer cell culturing, Mat. Sci. Eng. C 71 (2017) 465–472.
-
S. Gorodzha, T. E. L. Douglas, S. K. Samal, R. Detsch, K. Cholewa-Kowalska, et al., High-resolution synchrotron X-ray analysis of bioglass-enriched hydrogels: X-Ray Analysis of Bioglass-Enriched Hydrogels, J. Biomed. Mat. Res. A 104 (2016) 1194–1201.
-
R. V. Chernozem, M. A. Surmeneva, B. Krause, T. Baumbach, V. P. Ignatov, et al., Hybrid biocomposites based on titania nanotubes and a hydroxyapatite coating deposited by RF-magnetron sputtering: Surface topography, structure, and mechanical properties, Appl. Surf. Sci. 426 (2017) 229–237.
-
N. Aghaallaei, F. Gruhl, C. Q. Schaefer, T. Wernet, V. Weinhardt, et al., Identification, visualization and clonal analysis of intestinal stem cells in fish, Development 143 (2016) 3470–3480.
-
M. Takamiya, F. Xu, H. Suhonen, V. Gourain, L. Yang, et al., Melanosomes in pigmented epithelia maintain eye lens transparency during zebrafish embryonic development, Sci. Rep. (Nature) 6 (2016) 25046.
-
S. Hahn, Y. Müller, R. Hofmann, J. Mossmann, O. Öktem, et al., Spectral Transfer from Phase to Intensity in Fresnel Diffraction, Phys. Rev. A 93 (2016) 053834.
-
J. Moosmann, A. Ershov, V. Altapova, T. Baumbach, M. S. Prasad, et al., X-ray phase-contrast in vivo microtomography probes new aspects of Xenopus gastrulation, Nature 497 (2013) 374–377.
-
J. Moosmann, A. Ershov, V. Weinhardt, T. Baumbach, M. S. Prasad, et al., Time-lapse X-ray phase-contrast microtomography for in vivo imaging and analysis of morphogenesis, Nature Prot. 9 (2014) 294–304.
-
T. Nawy, Imaging: Embryos under the X-ray, Nature Meth. 10 (2013) 603 – 603.
-
T. van de Kamp, P. Vagovic, T. Baumbach, and A. Riedel, A Biological Screw in a Beetle’s Leg, Science 333 (2011) 52–52.
-
T. dos Santos Rolo, A. Ershov, T. van de Kamp, and T. Baumbach, In vivo X-ray cine-tomography for tracking morphological dynamics, Proc. Natl. Acad. Sci. 111 (2014) 3921–3926.
-
T. van de Kamp, T. dos Santos Rolo, P. Vagovic, T. Baumbach, and A. Riedel, Three-Dimensional Reconstructions Come to Life - Interactive 3D PDF Animations in Functional Morphology, PLoS ONE 9 (2014) e102355.
-
N. C. Wulff, T. van de Kamp, T. dos Santos Rolo, T. Baumbach, and G. U. C. Lehmann, Copulatory courtship by internal genitalia in bushcrickets, Sci. Rep. (Nature) 7 (2017) 42345.
-
A. H. Schwermann, T. dos Santos Rolo, M. S. Caterino, G. Bechly, H. Schmied, et al., Preservation of three-dimensional anatomy in phosphatized fossil arthropods enriches evolutionary inference, eLife 5 (2016) e12129.
-
T. van de Kamp, A. H. Schwermann, T. dos Santos Rolo, P. D. Lösel, T. Engler et al., Parasitoid biology preserved in mineralized fossils, Nature Comm. 9 (2018): 3325.
